From 2012.igem.org
I. Check Plates for Growth
RESULTS:
Growth on positive controls was a large number of colonies but not a lawn as expected. No growth on any of the Tetracycline plates.
II. Transform WT ADP1 using natural competence
- Inoculate 20 μL of the O/N cultures from yesterday into 300 μL of LB for 4 tubes as follows:
| Number | Name
|
| 1 | ADP1 (-)
|
| 2 | ADP1 +pBAV1K
|
| 3 | ADP1 +1:1 ligation
|
| 4 | ADP1 +1:3 ligation
|
- Incubate with shaking on its side (using tape to keep the tube in place) at 30 oC for 2.5 hours
- Add 2.5 μL of ~200 ng pBAV1K/μL sample for ADP1+pBAV1K, and 2.5 μL of sterile H2O for ADP1 (-)
- Add whatever was left from the ligation mixtures to the two ligation tubes (4-8 uL)
- Incubate with shaking as above for 2 hours
- For each sample, add 150 uL of cells to appropriate plate as follows using glass beads
| Sample | Plate
|
| Negative Control 1 (-) | LB + Kan50
|
| Negative Control 2 (-) | LB + Tet20
|
| pBAV1K | LB + Kan50
|
| pBAV1K Positive Control | LB
|
| 1:1 Ligation | LB + Tet20
|
| 1:1 Ligation Positive Control | LB
|
| 1:3 Ligation | LB + Tet20
|
| 1:3 Ligation Positive Control | LB
|
- Incubate at 34 oC until colonies form
III. Digest pBAV1K-T5-gfp Plasmid and Cassette with XbaI and PstI, and CIP for pBAV1K
NOTE: The plasmid digestion was started an hour before the cassette digestion so that the two digestions would end at the same time.
- 30.5 uL water
- 10 uL pBAV1K-T5-gfp DNA (~2 ug at 200 ng/uL)
- 0.5 uL 100x BSA
- 5 uL 10 NEB Buffer 3
- 2 uL XbaI
- 2 uL PstI
- Digest for 4 hours at 37 oC, then add 0.5 uL of CIP
- Incubate for another hour at 37 oC
- 40.5 uL cassette DNA (~1.8 ug at 45 ng/uL)
- 0.5 uL 100x BSA
- 5 uL 10 NEB Buffer 3
- 2 uL XbaI
- 2 uL PstI
- Digest for 4 hours at 37 oC
IV. PCR for More Cassette
- Mix according to the following table
| | Negative Control | Reactions (x4)
|
| Reagent | (uL) | (uL)
|
| water | 17.25 | 16.25
|
| 5x Fidelity Buffer | 5 | 5
|
| 10 mM dNTP mix | 0.75 | 0.75
|
| 10 uM primer (F) | 0.75 (pBf) | 0.75 (pBf)
|
| 10 uM primer (R) | 0.75 (p2Br) | 0.75 (pBr)
|
| Template ("Cassette 1") | - | 1
|
| KAPA HiFi polymerase | 0.5 | 0.5
|
| Step | Temp (oC) | Time
|
| 1 | 94 | 5 m
|
| 2 | 98 | 20 s
|
| 3 | 63 | 15 s
|
| 4 | 72 | 5 m
|
| 5 | GOTO 2 | 34x
|
| 6 | 72 | 5 m
|
| 7 | 4 | ∞
|
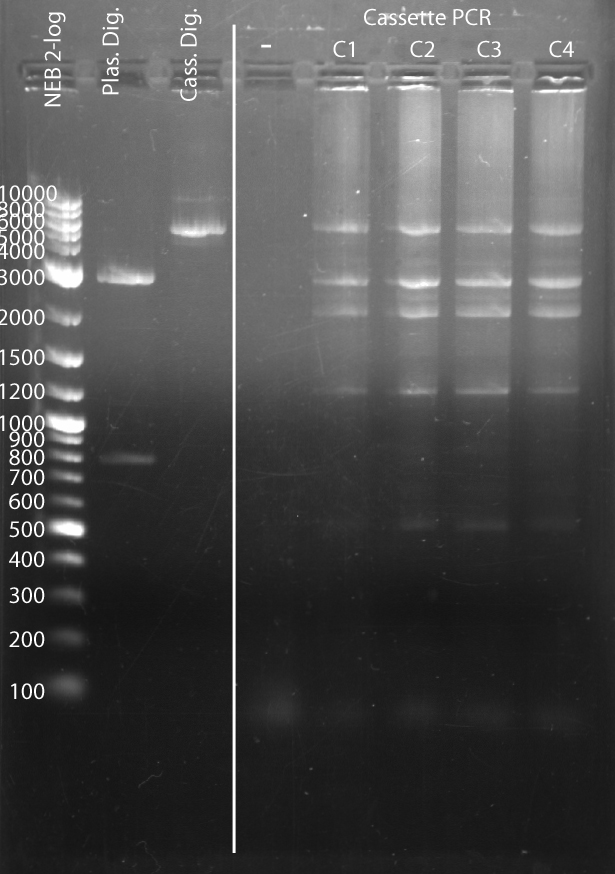
V. AGE of Digestion and PCR Samples
- Measure out 0.40660 g of agarose and add to a 250 mL E-flask
- Add 50 mL of 0.5x TBE buffer and swirl to mix (0.8% gel)
- Cover flask with a Kimwipe and microwave for ~1 minute until clear
- Allow to cool to ~60 oC and pour into beaker for ethidium bromide (EtBr)
- Add 1 uL of EtBr stock to agarose and swirl to mix
- Immediately pour gel into tray with combs
- Allow to solidify, then remove the comb and tape and place the tray in the gel box with the wells closer to the black electrode
- Make sure it is submerged in TBE buffer
- Mix samples as drops on a piece of parafilm
-
| NEB 2-log (Invtrogen 1 kb plus) | Sample
|
| *4 uL pre-mixed | *6.3 uL dH2O
*1.7 uL 6x Loading Dye
*2.0 uL DNA
|
- Load ~10 uL on gel according to the following chart (except ladder which is 4 uL):
| Lane 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8
|
| 2-log ladder | Plasmid Digestion | Cassette Digestion | (-) | C1 | C2 | C3 | C4
|
- Store DNA at 4 oC
- Unusual well formation in some cases
- Close gel box and turn on power pack
- Run gel at 30 V for ~50 min to stack gel
- Run gel at 75 V until the markers have reached ~3/4 of gel
-
Source: Media:Srk_2012-08-14_19hr_34min.tif
| Feature | Expected Size (bp)
|
| Uncut Cassette | 5152
|
| Cut Cassette | 5144
|
| Uncut Plasmid | 3653 (circular)
|
| Cut Plasmid | 2807
|
| GFP fragment | 846
|
- Measure out 0.40460 g of agarose and add to a 250 mL E-flask
- Add 50 mL of 0.5x TBE buffer and swirl to mix (0.8% gel)
- Cover flask with a Kimwipe and microwave for ~1 minute until clear
- Allow to cool to ~60 oC
- Immediately pour gel into tray with comb
- Allow to solidify, then remove the comb and tape and place the tray in the gel box with the wells closer to the black electrode
- Make sure it is submerged in fresh TBE buffer
- Add 9.6 uL of Loading Dye to each of the ~48 uL samples and mix.
- Load on gel according to the following chart:
| Lane 1 | 2 | 3 | 4 | 5 | 6 | 7
|
| NEB 2-log ladder (4 uL, premixed) | Plasmid Digestion (~9.6 uL) | Plasmid Digestion (~48 ul) | empty | NEB 2-log | Cassette Digestion (~9.6 uL) | Cassette Digestion (~48 ul)
|
- Close gel box and turn on power pack
- Run gel at 30 V for 20 min
- Run gel at 75 V until the markers have reached 3/4 of gel
- Remove gel from box and cut along lanes to separate the 9.6 uL lanes from the 48 uL lanes.
- Return 48 uL lanes to the gel box
- Place 9.6 uL lanes in 200 mL of 0.5 ug/mL EtBr solution for 15 minutes
- View 10 uL lane under UV to identify band of interest
- Use a clean razor blade to nick gel where the band is
- Match 48 uL lanes to the 9.6 uL lanes
- Using the 9.6 uL lane as a guide, cut out the band from the 48 uL lane
- Weigh each gel slice in a colorless 15 mL conical tube:
| Plasmid Digestion | Cassette Digestion
|
| 378 mg | 399 mg
|
- Add 3 volumes Buffer QG to 1 volume of gel
| Plasmid Digestion | Cassette Digestion
|
| 1134 uL | 1197 uL
|
- Incubate in 50 oC water bath for 12 minutes until all gel has dissolved. Vortex to help mix.
- Note: The solution was yellow, OK to proceed
- Add 1 volume 100% isopropanol to samples and mix by inversion
| Plasmid Digestion | Cassette Digestion
|
| 378 uL | 399 uL
|
- Apply each sample to a QIAquick column 800 uL at a time. Centrifuge for 1 minute at 13000 rpm, discard flow-through, and repeat until all of the sample has passed through the column
- Add 500 uL of Buffer QG to the QIAquick columns and centrifuge for 1 minute, discard flow-through
- Add 750 uL of Buffer PE to the QIAquick column and incubate on the bench for 5 minutes
- Centrifuge for 1 min and discard flow through
- Centrifuge again for 1 minute
- Transfer the columns to clean microcentrifuge tubes
- Elute DNA by adding 30 uL of sterile water to the center of each column and let stand for 4 minutes
 "
"